Innovative new research has revealed that the activity of different versions of genes expressed in the brain is associated with the accumulation of the protein tau, which is a hallmark of Alzheimer’s disease.
The University of Exeter Medical School research used new ‘long-read’ sequencing technology to progress understanding of how genes are expressed in the brain, revealing potential new targets for drug development to treat dementia.
We have approximately 20,000 genes encoded in our DNA sequence, but each gene can be expressed in many different versions – or isoforms – that are generated by a process called ‘alternative splicing’. This process can produce many isoforms of an expressed gene by sticking together different parts of the coding sequence in different combinations.
In a study published in Nature Communications and funded by Alzheimer’s Research UK, the Medical Research Council and the National Institute for Health and Care Research (NIHR) Exeter Biomedical Research Centre, the Exeter team used long-read genetic sequencing, combined with new data analysis tools they created. The new technique can read greater and more complex sequences of genetic material in one go than ever before. This allowed the team to map out the diversity of isoforms in the brains of mice adapted to carry a human mutant form of the protein tau, in the greatest detail to date.
Methodological Overview
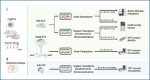
A An overview of the experimental approach used to profile isoform diversity in (i) the rTg4510 mouse model using short-read RNA-Seq (Illumina), long-read whole transcriptome and targeted Iso-Seq (PacBio) and long-read nanopore cDNA sequencing (ONT) from bulk cortex tissue and FANS-isolated nuclei populations and (ii) post-mortem human cortex tissue from individuals with high and low AD neuropathology using long-read targeted nanopore cDNA sequencing (ONT). B An overview of the bioinformatics pipeline used to process, align, quantify, annotate and characterize full-length transcript reads. Further details are provided in Methods. C An overview of the FICLE bioinformatics tool developed as part of this work to comprehensively characterize and visualize targeted long-read RNA sequencing data. AD Alzheimer’s disease, AS alternative splicing, CCS circular consequence sequence, CTX cortex, ECX entorhinal cortex, FANS fluorescence-activated nuclei sorting, FL full-length, NeuN neuronal nuclear protein, HQ high-quality, LR long-read, ONT Oxford Nanopore Technologies, PacBio Pacific Biosciences, WT wild-type mice, TG rTg4510 transgenic mice.
As well as finding hundreds of new isoforms of genes known to play a causal role in Alzheimer’s disease, they found specific isoforms that were associated with the accumulation of tau. They were then able to show that many of the isoforms found in mice were differentially expressed in tissue from human donors who had died with Alzheimer’s disease, showing the discoveries are relevant in the human brain. This work was supported by a research grant from Alzheimer’s Research UK.
The research builds on the team’s previous work, which found thousands of new isoforms in the brain linked to a wide range of disease.
Professor Jonathan Mill, of the University of Exeter Medical School, is senior author on the paper, and said: “This is incredibly exciting research which gives us far greater detail on how genes involved in dementia are expressed in the brain and reveals potential new drug targets for treating Alzheimer’s disease that could one day disable the mechanism by which the tau accumulates. The more we learn about the way Alzheimer’s disease genes are expressed in the brain, the more we realise how much more there is to discover, and this paper is a significant step along that path.”
Lead author Dr Szi Kay Leung, at the University of Exeter Medical School, said: “Advances in technology are allowing us to map out molecular changes in the brain in greater detail than ever before. Through that we’re learning that the complex pathway of gene expression is key in the development of Alzheimer’s and could also hold the key to new treatments. We’ve made our analysis pipeline available as a resource to the community to stimulate further research in this area.”
Source – University of Exeter
Leung SK, Bamford RA, Jeffries AR et al. (2024) Long-read transcript sequencing identifies differential isoform expression in the entorhinal cortex in a transgenic model of tau pathology. Nat Commun [Epub ahead of print]. [article]
