Scientists have developed incredible tools to study how genes are expressed in individual cells, giving us insight into the complex workings of tissues. Two key technologies are single-cell RNA sequencing (scRNA-seq) and spatial transcriptomics (ST). However, one big challenge has been combining these two approaches to understand not only what genes are active but where they are active in a tissue. This is where SELF-Former comes in—a new tool developed at the City University of Hong Kong that offers an innovative way to reconstruct spatial information from scRNA-seq data.
scRNA-seq analyzes gene activity in individual cells, helping researchers understand what each cell is doing. But it loses the spatial context, meaning it tells us which genes are active but not where the cells are located in the tissue. On the other hand, spatial transcriptomics (ST) captures both the gene activity and the location of cells within tissues, but it’s more challenging to perform. The trick is to align scRNA-seq data with ST data—essentially mapping gene activity back onto a tissue like putting puzzle pieces in the right places. But this is easier said than done. Batch effects (technical variations between datasets) and the need to filter out irrelevant gene information can make this task difficult.
SELF-Former is a powerful framework that tackles the challenge of aligning scRNA-seq data with spatial information. It uses transformer-based technology (similar to what powers modern AI) to analyze gene expression on multiple levels. SELF-Former learns patterns in gene activity to figure out which ones are relevant for spatial reconstruction and ensures the reconstructed data maintains the correct spatial relationships between genes, mapping them to the right locations. One of its standout features is the gene filtration module, which removes less informative genes to improve accuracy.
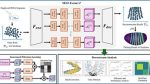
The flowchart of proposed SELF-Former F
We take a random gene from the drosophila as an example: firstly, high-throughput expression data for the gene is extracted from single-cell resolution, as shown in the left image which lacks spatial information. Secondly, the scRNA-seq data is encoded from Fenc, primarily implemented as a self-attention transformer block. The bottom left corner illustrates an overview of gene-wise filtration learning. Next, features are aggregated along multi-scale self-attention blocks to obtain spatially resolved predictions of ST data. The aggregated representations are then processed through the decoder Fdec. The decoder reconstructs the ST data from these multi-scale features. The final output is the reconstructed ST data, which includes spatial locations for each gene expression, allowing for spatial plotting and further downstream analysis.
The framework has been tested on four benchmark datasets, showing impressive results. It successfully recovers spatial information with high accuracy, reduces batch effects between datasets for improved consistency, and selects key genes that refine spatial mapping. These capabilities make SELF-Former a reliable tool for reconstructing spatial maps from single-cell data, providing researchers with a deeper understanding of how cells behave in their natural environments.
By accurately mapping gene expression to its location within tissues, SELF-Former opens up exciting possibilities. Researchers can better understand disease mechanisms by seeing how different cells interact within a tumor or damaged tissue. It could also help develop targeted treatments by identifying specific cell types driving disease and improve diagnostics by detecting gene patterns associated with particular conditions.
SELF-Former offers an innovative solution to one of the toughest challenges in biology: linking gene expression to spatial information. This advancement brings us closer to understanding how cells function within tissues and paves the way for breakthroughs in personalized medicine and disease research.
Availability – The implementation of SELF-Former is available at [https://github.com/bravotty/SELF-Former].
