RNA-sequencing has become a cornerstone in the study of gene expression, offering insights beyond mere mRNA transcript abundances. One area of increasing interest is alternative splicing, a process that allows a single gene to produce multiple transcript variants and thus protein isoforms with potentially different functions. In chronic lymphocytic leukemia (CLL), mutations in the splicing factor gene SF3B1 are well-documented, yet broader implications of SF3B1 mutations are underexplored.
Functional impact of SF3B1 mutations on the ncBAF chromatin remodeling complex through altered splicing
In a new study recently published in Leukemia, researchers from the Clinical Genetics research group at the Department of Molecular Medicine and Surgery conducted an in-depth analysis of SF3B1 mutations in CLL. The team, led by Dr. Richard Rosenquist Brandell, Professor of Clinical Genetics, focused on a specific subgroup of patients with CLL – those with a clinically aggressive form of the disease where SF3B1 mutations are found in up to 50%. Utilizing RNA-sequencing of samples from 35 patients, the team identified significant splicing events associated with these mutations, revealing multiple splicing events affecting the non-canonical BAF (ncBAF) chromatin remodeling complex, an important epigenetic regulator that controls the accessibility of DNA and influences gene expression by altering chromatin structure. This included splicing events in the ncBAF complex subunit BRD9 and eight additional ncBAF interactors. The findings were further validated through long-read RNA-sequencing and extended RNA-sequencing analysis of samples from 139 patients with CLL, solidifying the association between SF3B1 mutations and these splicing events.
Key findings: The role of BRD9 in CLL
This study is particularly notable for its emphasis on the ncBAF complex, an area that has so far received limited attention in CLL research. One of the study’s most intriguing findings is the identification of an alternative BRD9 splice variant that produced a protein isoform with an altered C-terminal structure. This isoform exhibited enhanced interaction with the ncBAF complex while showing reduced binding to other important proteins including SPEN, BRCA2, and CHD9.
“This is a significant step forward in our understanding of CLL pathobiology,” explained Dr. Rosenquist Brandell. “SF3B1 mutations have long been linked to poor prognosis in CLL. Our study highlights a novel pathobiological mechanism by which the spliceosome dysregulation caused by SF3B1 mutations alters the ncBAF complex’s interaction network, potentially contributing to the aggressive nature of SF3B1-mutated CLL.”
Long-read RNA-sequencing validates the predicted alternative transcriptsrelated to the ncBAF complex in SF3B1-mutated CLL
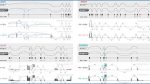
Sashimi plots illustrating the identified ASEs and alternative splicing patterns in four genes that encode ncBAF complex-related proteins in SF3B1WT versus SF3B1MUT CLL; ZEB1 and BRD9 exhibit the top two ASEs in the SE category, while DLST and SERBP1 showcase the top two ASEs in the A3SS category. Sashimi plots for other ncBAF complex-related genes, PLSCR1, TENT4B, CXXC1, DCAF16, and UBP1, are presented in Supplementary Fig. 3. For each gene, the top two sashimi plots within the gray box illustrate the predicted splice variants in SF3B1WT versus SF3B1MUT CLL. The colored arc highlights the primary ASE, while lighter arcs represent additional ASEs if present. The gene map indicates the relative location and order of the detected exons in relation to the sequencing results. For each corresponding gene, the lower two sashimi plots show the coverage and splice junction count data from the aligned long-read RNA-seq data from an SF3B1WT case (RS24) and an SF3B1MUT case (RS55), both belonging to subset #2 CLL. The direction of the genes is arranged from left to right. WT: wildtype; MUT: mutated; PSI: percent spliced in.
Research highlights the need for advanced splicing tools
The study also highlights the importance of alternative splicing in cancer research.
Blaz Oder, a PhD student and joint first author, noted, “Most RNA-sequencing studies in CLL focus predominantly on gene expression changes, but our work demonstrates the importance of alternative splicing – a dimension often overlooked but important in cancer biology, especially in the context of splicing factor mutations like SF3B1.”
The discovery of these splicing events underscores the need for more specialized research tools to study alternative splicing in cancer.
“To fully understand complex diseases like CLL, we need to develop disease-specific splicing databases and expand public datasets to include the splicing dimension,” said Dr. Daniel Hägerstrand, joint first author of the study. “Our work, which also utilized publicly available datasets, underscores the potential in already published data and the importance of further exploring these valuable resources.”
Implications for future CLL research
The team’s findings have important implications for future research, particularly highlighting the specific role of BRD9 in CLL. The discovery of the novel BRD9 isoform and the altered interactions of the ncBAF chromatin remodeling complex suggest that BRD9 could be a potential therapeutic target for SF3B1-mutated CLL. The ncBAF complex, and BRD9 in particular, is currently being investigated as a therapeutic target in several early clinical trials for other cancer types. The study demonstrated that BRD9 depletion sensitizes both cell lines and primary CLL cells, though further investigation is needed to better understand the therapeutic potential and how best to leverage these findings. More broadly, the research emphasizes the significance of SF3B1 mutations in the aggressive behavior of CLL and draws attention to the importance of alternative splicing in cancer biology more generally.
Source – Karolinska Institutet
