Spatial transcriptomics (ST) is reshaping how scientists study gene expression in tissues. Traditional methods like immunostaining and RNA in situ hybridization focus on a limited number of genes, which restricts researchers’ ability to get a complete picture of what’s happening in a tissue sample at the molecular level. ST, however, allows scientists to explore the entire set of genes being expressed in a tissue section, revealing an enormous amount of data from just a single histological slide. But, as powerful as ST is, it’s not without challenges: low resolution, limited gene range, high costs, and complex equipment requirements have slowed down its application in many labs.
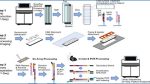
A new technology developed at the University of Michigan Medical School called Seq-Scope offers a way around these hurdles by repurposing the widely-used Illumina sequencing platform to analyze gene expression in tissues at high resolution. Instead of just identifying a few genes at a time, Seq-Scope can map many genes across multiple tissue sections on a 7mm x 7mm surface. This means that researchers can gain detailed insights into gene activity across different tissue areas, allowing for both high-resolution images and comprehensive gene expression data.
Here’s how Seq-Scope works: using an Illumina NovaSeq 6000 sequencing flow cell, researchers can prepare frozen tissue sections for both imaging and sequencing. This allows for easy merging of visual data from tissue structure (histology) with gene expression information (transcriptomics). By integrating these two types of data, scientists can see exactly where in a tissue certain genes are active, which is incredibly useful for studying diseases and tissue function.
Experimental Procedures
The current experimental procedures are divided into three major steps: Step 1 produces a spatially barcoded array, Step 2 attaches the tissue to the array, performs tissue imaging, and prepares the tissue for library construction, and Step 3 captures the tissue transcriptome through reverse transcription and processes the spatially barcoded cDNA into a next-generation sequencing library
The Seq-Scope method is also equipped with a “segmentation-free” analysis approach. Traditional ST methods often divide (or segment) tissue into specific regions, which can limit the resolution of the data. With Seq-Scope, scientists don’t need to rely on segmentation. Instead, they can perform spatial analysis directly at the submicrometer scale, allowing for a closer look at how genes are expressed in very tiny areas within the tissue. This adds another layer of detail that is difficult to achieve with older methods.
Overall, Seq-Scope is a powerful tool that brings high-resolution and comprehensive gene expression mapping to molecular biology and histology research. By making it easier to obtain high-quality, spatially detailed gene expression data across diverse tissues, Seq-Scope opens up new possibilities for understanding the molecular basis of diseases and biological processes with unprecedented clarity.
