While not everything in this world improves with age, technologies tend to get better and cheaper over time. This is good news for the sciences. Moore’s oft-cited law describes how the number of transistors that can be squeezed onto an integrated circuit doubles approximately every two years, with attendant improvements in processing power and little effect on cost.
The field of biology has benefited from a similar trend related to sequencing, which is the process of determining the sequence of molecules like DNA and RNA that encode genetic information. When it finished sequencing a human genome for the first time ever in 2003, the Human Genome Project did so to the tune of $3 billion. Today, the same thing can be done for a few hundred dollars.
“One of the really important things that has happened in the past decade is that DNA sequencing has become a commodity,” says Eduardo da Veiga Beltrame, assistant professor of computational biology at the Mohamed bin Zayed University of Artificial Intelligence. “The cheaper sequencing is, the more questions scientists can ask, and we can do it at scale. Today, if we can turn the readout of a biology experiment into a sequencing problem, we can collect immense amounts of data.”
Beltrame recently joined MBZUAI’s department of computational biology and is developing methods and tools for analyzing the huge amount of data that scientists are producing with these increasingly affordable sequencing technologies.
The cost of sequencing a human genome has decreased even faster than Moore’s Law over the past two decades. Source: Wetterstrand KA. DNA Sequencing Costs: Data from the NHGRI Genome Sequencing Program (GSP) Available at: www.genome.gov/sequencingcostsdata. Accessed 17 September 2024.
The codes: DNA, RNA and proteins
The genome is the complete set of genetic information in an organism, and dictates how it develops, grows and ages. But scientists can also sequence RNA, which is derived from DNA through a process known as transcription and encodes the information of what genetic programs are active in the cell. RNA is used by cells to make proteins through a process known as translation. Because RNA encodes information about what proteins the cell is making and what biological programs are active, measuring RNA is very informative. While the DNA of a particular organism is generally the same across cell types and at different moments in time, RNA varies according to cell type and changes frequently. This provides information about how genes are expressed.
“If you want to look at what a cell is doing right now, you need to look at the RNA,” Beltrame says.
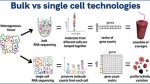
Much of Beltrame’s recent work relates to a powerful technique called single-cell RNA sequencing, which measures the RNA inside thousands of individual cells, providing insight into the molecular state of each cell. Scientists have been sequencing RNA for years, but previous approaches of “bulk RNA sequencing” mixed all the cells together, with the result being an RNA profile that would be an average of all the cells contained in the sample.
Single-cell RNA sequencing preserves RNA molecule count from each cell type, providing greater detail related to how genes are expressed in different cell types. Figure courtesy of Eduardo Beltrame.
If traditional RNA sequencing is like drinking a fruit smoothie, single-cell RNA sequencing is like eating a blueberry, Beltrame explains. The high resolution that single-cell RNA sequencing provides is particularly important for studying the underlying causes of disease, or identifying disease subtypes, as small numbers of cells often play a role in disease.
“In a sample of 10,000 cells, you may have 10 or 20 cells of a rare cell type that determine an important biological process,” Beltrame says. “If you were to do an average of all the cells, you would lose the signal of the rare cell types.”
Personalized medicine
In addition to improving our fundamental understanding of biology, the study of RNA, or transcriptomics as it’s known, has practical implications for personalized medicine, an approach to treating disease that considers the individual biological and molecular variability of people.
Two people may have what appears to be the same disease based on observed symptoms, but the molecular causes of the ailments could be different, Beltrame explains. Most autoimmune diseases are families of complex diseases and sub-diseases, with similar symptoms but different underlying mechanisms. Inflammatory bowel disease (IBD), for example, is not a single disease, but a group of inflammatory conditions, of which the most widely known are ulcerative colitis and Crohn’s disease — each with different causes, molecular mechanisms and treatments.
Being able to create precision diagnostics is essential for advancing precision medicine, and the high-resolution data yielded by single-cell RNA sequencing holds the promise of allowing scientists to gain a deeper understanding of the causes of disease and choose treatments that are best suited for a patient’s unique biology.
Beltrame believes that in the future we will benefit from frequent and data-rich measurements in the form of what he calls “routine single-cell omics,” which will provide people with the ability to analyze their health at home. People could test themselves twice a day — morning and evening, due to the strong influence of the circadian rhythm on cell function — and generate a detailed profile of their overall health. This stands in contrast to how diagnostics work today, where data-rich measurements like blood tests are taken rarely, and less revealing information, like blood sugar levels, can be gathered more frequently with the help of a device like a continuous glucose monitor.
Beltrame’s vision of routine single-cell omics and multiomics (red) will become possible as sequencing costs continue to decrease. Figure courtesy of Eduardo Beltrame.
“These rich measurements will become cheaper and easier to perform, and we are going to have 1,000 times more data than we do currently,” Beltrame says. “But to enable personalized medicine, we need to develop tools that can analyze data at this huge scale, and machine learning is the only paradigm that I have seen that holds this promise.”
A new era for medicine
One of the challenges to developing tools and methods for analyzing biological data is that it’s difficult to determine a “ground truth” in a molecular dataset.
“It’s not like an image that can be annotated as a cat rather than a dog. Because molecular biology is so complex, we don’t have an intuitive picture of it, and making sure we are on the right track requires a lot of biological literature deep dives that ensure the results we have are real, and not noise or experimental errors,” Beltrame says.
He believes that MBZUAI is the best place to pursue this line of research and develop new methods and tools for analyzing biological data because of the expertise of scientists at the university. This “critical mass” of knowledge at MBZUAI will only expand as more researchers with expertise in biology join the computational biology department, he says.
Beltrame’s vision of routine single-cell omics has the potential to transform how we diagnose and treat diseases. New machine learning tools will help researchers decode this flood of biological data and better understand the molecular processes that influence individual health. Physicians will be able to tailor treatments based on individual biology. These innovations may even empower people to take greater control of their own health with insights from frequent, data-rich diagnostics, marking a shift from occasional medical snapshots generated during annual doctors’ visits. In the meantime, there remains a lot of work to be done to realize this vision.
As Beltrame puts it, “Single-cell omics will revolutionize personalized medicine. We are going to have diagnostics for everything, even for mental health conditions like depression and bipolar disorder. Today for these diseases, we have to rely on observations of patients, but even mental health disorders have molecular correlates.”
Source – Mohamed bin Zayed University of Artificial Intelligence
