In a landmark study published today in Science Advances, Dr. Shai Bel and his research team at the Azrieli Faculty of Medicine of Bar-Ilan University have uncovered crucial insights into how antibiotic use increases the risk of inflammatory bowel disease (IBD).
Using sophisticated techniques like RNA sequencing and machine learning, the team highlights the need for new treatments that prevent these detrimental effects.
The study demonstrates that antibiotics interfere with the protective mucus layer in the intestine, a discovery that could reshape our understanding of antibiotic effects and IBD development.
IBD, which includes Crohn’s disease and ulcerative colitis, affects approximately 1% of the global population. This debilitating condition is marked by the erosion of the mucosal layer that serves as a critical barrier between the gut microbiome and the immune system. Despite ongoing research, the exact causes of IBD remain unclear. However, previous studies have indicated a link between antibiotic use and an increased risk of developing IBD.
Dr. Bel’s latest research sheds new light on this association.
“We have discovered that antibiotic use actually damages the protective mucus layer that separates the immune system in the gut from the microbiome,” says Dr. Bel. His team’s study reveals that antibiotics, whether administered orally or via injection, disrupt this vital mucus layer, facilitating bacterial penetration and increasing the risk of gut inflammation.
Utilizing advanced techniques such as RNA sequencing, machine learning, and mucus secretion measurement, the researchers examined the effects of antibiotics using mice models. The study found that antibiotics hinder the secretion of protective mucus, leading to bacterial infiltration, systemic bacterial antigen replication, and ulcer development — hallmarks of IBD.
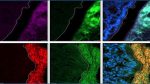
Vancomycin-induced changes to the gut microbiota cannot explaintreatment impact on the mucus barrier and gut transcription
(A) FISH images of colonic tissues from SPF mice treated with vancomycin as indicated or GF mice that received an FMT from vancomycin-treated mice and stained with the indicated probes. The dashed white lines mark the edge of the host epithelium. Scale bars, 20 μm. (B) Quantification of distance between luminal Gammaproteobacteria and host epithelium as in (A). (C) Quantification of the ratio of the distances between Clostridia or Gammaproteobacteria and the host epithelium as in (A). (D) PCA plot of colonic transcriptional profiles of mice treated as indicated. (E) A confusion matrix depicting the percentage of predictions for each category against the true classifications in a four-way Random Forest classification task. Diagonal entries represent the accuracy of predictions for each category (true positives), while off-diagonal entries indicate the model’s misclassifications. (F) Heatmap showing transcriptional changes of the top 200 genes that distinguish between the treatment groups on the basis of the classifier in (E). Each column represents a mouse. (G) Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways of transcripts which are uniquely altered in vancomycin-treated mice (donors) as plotted in (F) (in the black box). (B to D) Each dot represents a mouse. (B and C) One-way ANOVA. At least 25 measurements per mouse were taken. *P < 0.05; **P < 0.01. PO, per os.
One of the most striking findings of the study is that antibiotics’ impact on the mucus barrier is not due to alterations in the microbiome but rather affects the cells in the intestinal wall responsible for mucus production.
“This finding shatters the paradigm that antibiotics harm only bacteria and not our own cells,” Dr. Bel explains.
As a next step, the research team plans to explore potential treatments that could mitigate the adverse effects of antibiotics on mucus secretion. These findings not only advance our understanding of IBD but also underscore the need for careful consideration of antibiotic use and its broader implications for gut health.
Source – Bar-Ilan University
