Transfer RNAs (tRNAs) are critical molecules involved in protein synthesis, but they can be tough to analyze. This is mainly because tRNAs are highly modified, making traditional methods like reverse transcription and RNA sequencing (RNAseq) unreliable for accurately quantifying them. However, researchers at Johannes Gutenberg University Mainz have now developed a new approach that overcomes these challenges, offering a faster and more accurate way to study tRNA.
Why Traditional Methods Struggle
When scientists try to quantify tRNAs using RNAseq, they often encounter biases due to the multiple steps involved, especially during reverse transcription (converting RNA to DNA). These biases can lead to inaccurate results, making it difficult to get a clear picture of tRNA composition. To address this, scientists typically apply correction factors, but these add complexity and slow down the process.
The New Hybrid Approach
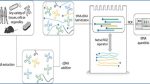
The new method developed by researchers bypasses these challenges. Instead of relying on reverse transcription, it uses a hybrid approach that combines hybridization-based techniques with deep sequencing. This method simplifies the process by transferring information about the tRNA pool into a cDNA mixture in one step. The only enzymatic process required is a final barcoding PCR, which labels the samples for sequencing.
Key Benefits
Fast and Cost-effective: This approach is quicker and cheaper than traditional methods.
Accurate and Sensitive: It provides detailed information on the composition of tRNAs from very small amounts of total tRNA (even in femtomolar quantities).
High-throughput: It’s suitable for large-scale studies, including clinical samples.
Simple Data Analysis: The bioinformatics required to process the data is straightforward, making it accessible for broader applications.
Applications and Findings
This new technique has already been applied to various biological research areas, leading to novel discoveries. For example, researchers have used it to:
Quantify tRNA bound to polysomes (structures involved in protein synthesis).
Study tRNA modifications in knockout strains under stress conditions.
Analyze tRNA composition in brain tissues from Alzheimer’s patients.
Conclusion
This breakthrough method simplifies tRNA quantification, making it faster, more accurate, and easier to apply to different biological and clinical questions. It holds great potential for advancing our understanding of tRNAs in health and disease, opening the door to new discoveries, especially in conditions like neurodegenerative diseases.
