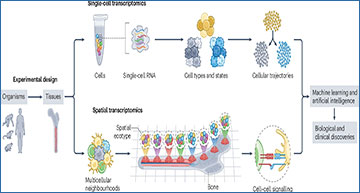
In recent years, scientists have made groundbreaking advances in understanding how individual cells function within the complex environments of our bodies. One key area of research is transcriptomics, which studies how genes are expressed in cells. Traditionally, transcriptomics has been done on groups of cells, but new technologies allow us to look at gene expression in single cells, providing a much clearer picture of cellular diversity and how cells behave in both healthy and diseased tissues. A team led by researchers at the Dana-Farber Cancer Institute and Stanford University discuss recent progress in sample processing, data integration, identification of subtle cell states, trajectory modelling, deconvolution and spatial analysis in single-cell transcriptomics
What is Single-Cell Transcriptomics?
Single-cell transcriptomics is like zooming in on individual cells to see which genes are active at any given moment. This technology has been revolutionary in identifying the unique characteristics of different cells within a tissue, revealing how even cells that look similar can have very different functions. By studying gene expression at the single-cell level, researchers have uncovered new insights into how cells develop, how they change in response to disease, and how diverse the environments within our tissues really are.
Key technical considerations in scRNA-seq
a, Example workflow of sample processing. Tissue is dissected from an organism and subjected to mechanical and then chemical digestion before single-cell capture and RNA sequencing. Time after death (‘post-mortem interval’), tissue-specific chemicals (for example, bile, bile acid, gastric contents and extracellular enzymes), and tissue-preservation protocols (for example, formalin-fixed, paraffin-embedded (FFPE)) are other variables that can affect the resulting cellular and transcriptome composition. b, Data integration methods aim to combine single cells from different samples into a unified low-dimensional embedding (top). Batch correction methods modify the high-dimensional gene expression matrix to preserve biological signal while removing noise from unwanted technical covariates (bottom). c, The production of cell atlases involves curating millions of single cells from hundreds of tissues and species to enable reference-based annotation of single-cell transcriptomic data and the discovery of new cell states and types. d, Identification of rare and subtle cells, such as Hoxb5-expressing haematopoietic stem cells (HSCs), with single-cell transcriptomics requires careful consideration of sample processing, gene selection, enrichment protocols and library preparation platforms: digestion and dissociation protocols can alter the composition of cells in the sample and gene expression programmes (top middle); several methods have been developed to optimize the selection of marker genes to identify rare and subtle cells (top right); prior knowledge of anatomic location and expression of cell surface markers can help enrich for desired cell populations with laser microscopy and flow cytometry, respectively (bottom left, middle); and the number of cells, number of genes and types of transcripts captured by different platforms (for example, droplet-based, plate-based and targeted transcriptomics) can influence the likelihood of detecting rare and subtle cell states (bottom right).
Introducing Spatial Transcriptomics
While single-cell transcriptomics is incredibly powerful, it doesn’t tell us everything. It can show us what’s happening inside individual cells, but not how those cells are interacting with their neighbors. That’s where spatial transcriptomics comes in. This new approach allows scientists to see where cells are located in relation to each other within a tissue and how they interact. For example, spatial transcriptomics can reveal how cells in a tumor communicate with each other, which could help in developing more effective cancer treatments.
Challenges and Innovations
As exciting as these technologies are, they come with challenges. Analyzing the vast amounts of data generated by single-cell and spatial transcriptomics requires sophisticated computational tools. Researchers are continually developing new methods to process, integrate, and interpret this data, which is crucial for identifying subtle differences in cell states and mapping out the complex networks of cells in tissues.
One area that’s seeing rapid advancement is the use of deep learning—an artificial intelligence technique—to analyze transcriptomics data. Deep learning models can help scientists make sense of the complex patterns in gene expression data, potentially leading to new discoveries in how cells function and interact.
Applications in Medicine
These cutting-edge tools are not just for basic research; they have real-world applications in medicine. In stem cell biology, single-cell and spatial transcriptomics are helping scientists understand how stem cells develop into different types of cells, which could lead to new regenerative therapies. In immunology, these technologies are being used to study how immune cells respond to infections and how they might be manipulated to fight diseases like cancer. And in tumor biology, spatial transcriptomics is revealing how cancer cells interact with their surrounding environment, which could lead to more targeted and effective treatments.
The Future of Cellular Research
Looking ahead, the future of single-cell and spatial transcriptomics is bright. As these technologies continue to evolve, they will likely become even more integrated into both research and clinical practice. Scientists hope to use these tools to develop more personalized treatments for diseases, tailored to the unique cellular makeup of each patient’s tissues.
In summary, single-cell and spatial transcriptomics are transforming our understanding of biology at the most fundamental level. By providing a detailed view of how individual cells function within their environments, these technologies are opening up new avenues for research and offering promising new strategies for treating diseases.
