Understanding the complex organization of mammalian genomes is crucial for comprehending how our genes function and are regulated. This organization isn’t just a simple sequence of DNA but involves a complex, three-dimensional (3D) architecture. Despite its importance, the relationship between this 3D structure and gene activity has remained somewhat mysterious due to technological limitations. Traditional methods could either look at genome organization or gene activity, but not both at the same time in the same cell.
This gap in understanding is being addressed by a new technique called GAGE-seq (Genome Architecture and Gene Expression by Sequencing) developed by researchers at Carnegie Mellon University. GAGE-seq is a groundbreaking method that can simultaneously examine the 3D structure of the genome and the gene activity within individual cells. This is like getting a detailed blueprint of a building and knowing what activities are happening in each room all at once.
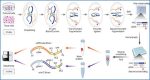
Overview and validation of GAGE-seq
a, Schematic representation of the GAGE-seq workflow detailing the simultaneous single-cell profiling of 3D genome architecture and gene expression. b–e, Validations demonstrating the specificity of GAGE-seq using mixed experiments with the human (K562) and mouse (NIH3T3). b,d, Scatter plots showing the collision level in the GAGE-seq scHi-C (b) and scRNA-seq (d) libraries, and histograms showing the binomial distribution of reads mapped to hg38 (top) and mm10 (right). c, Scatter plot showing the cis–trans ratio of scHi-C reads. The gray dashed line shows y = x, that is, the cis-to-trans ratio equals to 1. e, Scatter plot showing the well-separation of scHi-C and scRNA reads of valid cellular indices from that of empty indices. Green, mouse; orange, human; red, collisions; gray, empty indices.
The researchers applied GAGE-seq to cells from the mouse brain cortex and human bone marrow. In the mouse brain, they were able to see how the 3D organization of the genome influences which genes are turned on or off in different types of brain cells. This helped them understand how specific regulatory elements (parts of the genome that control gene activity) are linked to their target genes.
In human bone marrow cells, which are crucial for blood formation, GAGE-seq revealed some surprising findings. It showed that changes in the 3D organization of the genome and changes in gene activity don’t always happen in sync. This suggests that the relationship between genome structure and gene expression is quite complex and can vary over time within individual cells.
One particularly exciting aspect of this research is how GAGE-seq was combined with spatial transcriptomics data, which provides information on where genes are active within tissues. This allowed the researchers to see how 3D genome organization varies in different areas of the mouse cortex, adding another layer of understanding to how the genome functions in different cellular environments.
Overall, GAGE-seq is a powerful, cost-effective tool that opens up new possibilities for studying the genome at an unprecedented level of detail. It helps scientists explore how the 3D structure of the genome and gene activity are interconnected in various biological contexts, from brain function to blood cell formation. This could lead to a deeper understanding of many biological processes and diseases, paving the way for new therapeutic strategies.
Zhou T, Zhang R, Jia D, Doty RT, Munday AD, Gao D, Xin L, Abkowitz JL, Duan Z, Ma J. (2024) GAGE-seq concurrently profiles multiscale 3D genome organization and gene expression in single cells. Nat Genet [Epub ahead of print]. [abstract]
