RNA molecules play crucial roles in various processes, including gene expression, protein synthesis, and cellular function. One significant aspect of RNA research is understanding how modifications to RNA can influence these processes. One such modification is known as 5-methyluridine (m5U), and identifying where these modifications occur in RNA molecules is essential for grasping their overall function.
What Are RNA Modifications?
RNA modifications are chemical changes made to RNA molecules after they are formed. These modifications can alter how RNA behaves, affecting everything from how genes are expressed to how cells respond to environmental signals. m5U is one of these modifications, and it is believed to play a crucial role in maintaining the integrity, structure, and function of RNA. Therefore, scientists aim to locate and study these m5U sites to better understand their impact on biological processes.
Introducing GRUpred-m5U
To enhance our ability to detect m5U sites, a team led by researchers at Oakland University have developed a new tool called GRUpred-m5U. This innovative tool uses deep learning, a type of artificial intelligence, to analyze RNA data. It operates by utilizing complex algorithms that can learn patterns and make predictions based on large datasets.
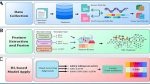
Overview of the m5U analysis workflow
(A) collection of the two types of datasets: the full transcript and mature mRNA, (B) feature extraction and fusion method to create a two-stage feature selection approach, (C) applying deep learning model on cross-validation evaluation method, (D) performance assessment with the evaluation metrics and finally discovering the active class from the datasets.
The researchers applied three different groups of descriptors to help the model understand RNA:
Nucleic Acid Composition: This refers to the basic building blocks of RNA.
Pseudo Nucleic Acid Composition: This involves a modified understanding of nucleic acids, considering certain characteristics that might influence their behavior.
Physicochemical Properties: These are the physical and chemical properties of RNA molecules, which can affect how they interact with other molecules in the cell.
To further enhance the model’s performance, five different feature extraction methods were combined, allowing the model to gather as much relevant information as possible from the RNA data.
Performance and Evaluation
The researchers tested GRUpred-m5U using deep learning techniques and compared its accuracy to existing methods. They conducted extensive evaluations through a process called 10-fold cross-validation, which ensures the model is reliable and not just a fluke. Remarkably, the GRUpred-m5U model achieved impressive accuracy rates of 98.41% and 96.70% on two different RNA datasets, surpassing all previously existing technologies in this area.
Additionally, the model underwent an evaluation using unsupervised machine learning techniques, like principal component analysis (PCA), which helped confirm its effectiveness in identifying m5U sites. The advanced design of GRUpred-m5U allows it to excel at recognizing complex patterns, making it a powerful tool for researchers in the field.
Implications for the Future
The successful development of GRUpred-m5U marks a significant advancement in RNA research. With its ability to accurately identify m5U modifications, this model opens up exciting possibilities for future applications in the biological industry. Researchers can utilize this tool to gain deeper insights into RNA biology, which could lead to breakthroughs in understanding diseases, developing new therapies, and advancing the overall field of molecular biology.
In summary, the GRUpred-m5U model is not just a technological achievement; it’s a step forward in unraveling the complexities of RNA modifications and their implications for health and disease. As this research continues to unfold, we can expect exciting developments that could revolutionize our understanding of cellular function and disease mechanisms.
