Molecular quantitative trait loci (molQTL) mapping is a powerful tool that helps scientists understand how genetic variations can affect the molecular processes inside cells, shedding light on the underlying causes of diseases and complex traits. One of the most common types of molQTL is expression quantitative trait loci (eQTL), which focuses on how certain genetic differences can influence gene expression—essentially how much or how little a gene is “turned on” in different individuals. These insights are crucial because changes in gene expression can impact everything from metabolism to immune responses, contributing to diseases like cancer, diabetes, and autoimmune disorders.
However, while molQTL studies can offer tremendous value, they are not always easy to conduct. The field involves many advanced tools and statistical methods, which can be daunting for researchers who lack a background in computational biology. This is particularly true for eQTL analysis, which requires processing large amounts of genetic and expression data to identify meaningful patterns and connections. To help make these studies more accessible, researchers at POSTECH have compiled a curated list of the most effective tools and software for analyzing eQTLs.
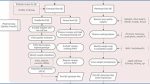
Framework diagram of eQTL mapping
A schematic diagram of the eQTL mapping process with commonly used tools for each step.
This guideline offers a starting point for scientists who may not be experts in programming or bioinformatics but still want to explore how genetic variations impact gene expression. With user-friendly tools and step-by-step approaches, researchers can better navigate this field and uncover important findings, potentially opening doors to new therapies and treatments for complex diseases. This guidance ensures that more people can participate in molQTL research, making it easier to bridge the gap between genetics and real-world medical applications.
