Colorectal cancer (CRC) is a complex disease that doesn’t affect every patient in the same way. As cancer cells evolve, they form different groups, or sub-clones, that can behave in varied ways—some may spread (metastasize) more easily, and others might respond differently to treatments. This diversity makes it difficult for doctors to predict which patients will experience metastasis, the spread of cancer to other parts of the body, which is a key factor in determining how serious the disease becomes.
To improve patient outcomes, researchers have been searching for reliable biomarkers, which are molecules that can indicate the likelihood of disease progression or treatment response. Historically, the focus has been on proteins, but recent studies are shifting attention to another class of molecules: non-coding RNAs. Unlike the RNA molecules that carry instructions for making proteins, these non-coding RNAs don’t directly make proteins, yet they still play essential roles in how cells function.
One specific type of non-coding RNA, known as long non-coding RNAs (lncRNAs), makes up the majority of RNA in the body. These lncRNAs are especially interesting because they are highly specific to certain tissues and types of cancer. This means they could serve as precise indicators of disease, especially in cancers like CRC, where understanding the behavior of cells is key to predicting how the cancer will progress.
In this study, scientists at the University of Otago used a technique called spatial transcriptomics to get a clearer picture of where these lncRNAs are located within the tumor tissue itself. This technique is more accurate than traditional methods like bulk RNA sequencing, which averages out RNA expression across many cells, because spatial transcriptomics can show what is happening at the level of individual cells and specific regions of the tumor.
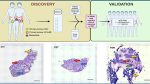
Spatial transcriptomics confirms the spatial architecture of cellular tissue components
A Overview of methodology for identification and characterisation of lncRNA markers with potential clinical relevance. B H&E staining of tumour sections used for Visium spatial gene expression profiling, with pathologist annotations. Left = P1T, Middle = P2T, Right = P2M. Red arrows denote areas with high nuclei density (malignant lesions). Blue arrows denote adjacent normal tissue. Yellow arrows denote necrosis. Green arrows denote desmoplastic stroma (dense, fibrous tissue with low cellularity). Scale bars = 1000 µm. C Unbiased clustering of spots for P1T, P2T and P2M using the FindClusters algorithm (Seurat v4), with assigned cell or tissue types based on transcriptomic profile. Missing spots = removed during the QC process. D Tumour purity analysis based on cell type clusters from C. Adjacent normal = “Colon epithelium” and “Colon epithelium in EMT”. Tumour = all other cell types. E Distribution of cell and tissue types in each tissue, calculated as the sum of spots associated with each cluster, per tissue. F Percentage of total spots associated with each tissue. Image created with Biorender.com.
By using this method, the researchers identified 301 lncRNAs that were associated with malignant (cancerous) regions of CRC tumors. To make sure these findings were accurate, they compared them with data from publicly available studies, and the results held up. They then conducted further testing using a technique called RNA–FISH (Fluorescence In Situ Hybridization) to see exactly where three specific lncRNAs—LINC01978, PLAC4, and LINC01303—were being expressed.
What they found was particularly exciting. These three lncRNAs were present in stage II tumors (a relatively early stage of CRC) but not in normal tissue. Moreover, they were even more abundant in metastatic tissues, which are the cancer cells that have spread to other parts of the body. This suggests that these lncRNAs could serve as early warning signs for patients who are at risk of their cancer spreading.
The discovery of these lncRNAs as potential biomarkers for predicting metastasis is a significant step forward in CRC research. By identifying these molecules early on, doctors could potentially assess a patient’s risk of metastasis before it happens, allowing for more tailored and timely treatments. This could improve the chances of successful intervention and help prevent the cancer from spreading to other parts of the body, ultimately leading to better patient outcomes.
This research highlights the growing importance of lncRNAs in cancer biology and their potential to serve as powerful tools for early detection and risk assessment in colorectal cancer. As our understanding of these molecules deepens, they could open up new avenues for diagnosis and treatment, offering hope to patients facing this challenging disease.
