Scientists have made significant strides in understanding the complex world of RNA using single-cell RNA sequencing (scRNA-seq). This technique allows researchers to study the RNA in individual cells, helping them to identify different cell types, their states, and how they change over time. However, traditional scRNA-seq mostly uses short-read sequencing, which isn’t detailed enough to fully capture the diversity of RNA molecules, known as RNA isoforms, within a single cell.
RNA isoforms are different versions of RNA produced from the same gene. Think of them as alternative recipes that can be made from the same cookbook. These different versions can result in slightly different proteins, which may have unique functions in the body. Understanding these isoforms is crucial for a deeper insight into how cells function, particularly in diseases like cancer.
scTaILoR-seq (Single-cell Targeted Isoform Long-Read Sequencing) is a new method that tackles this challenge. Developed by researchers at Genentech to improve the detection of RNA isoforms at the single-cell level, scTaILoR-seq uses a technique called hybridization capture to target over a thousand specific genes. This approach increases the amount of relevant RNA data that can be collected from each cell by 29 times compared to previous methods.
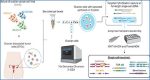
Overview of single-cell long-read targeted sequencing
Ovarian cell lines or dissociated tumor cells are processed using droplet-based single-cell RNA-seq 3’-Gene expression assay to obtain cDNA. Targeted enrichment is performed followed by nanopore sequencing, cell barcode (CB) and unique molecular identifier (UMI) assignment, and read alignment. Downstream analysis enables the measurement of isoforms, SNVs, allelic expression, TCR sequences and gene fusions at single-cell resolution.
To test their new technique, the researchers applied scTaILoR-seq to study ovarian cancer cells, both from lab-grown cell lines and from actual tumors. They successfully identified and quantified various RNA isoforms in 10,796 single cells. But they didn’t stop there—by using a process called long-read variant calling, they could also detect small genetic differences, known as single nucleotide variants (SNVs), within these RNA molecules. These SNVs can impact which RNA isoforms are produced and can differ from one cell population to another.
Moreover, the scTaILoR-seq method allowed the researchers to phase SNVs across transcripts. Phasing is like connecting the dots between genetic variations and their impact on RNA, enabling the scientists to observe how these variations lead to imbalances in RNA isoform production across different cells. This can be particularly important in understanding how certain cell populations within a tumor might respond differently to treatments.
In summary, scTaILoR-seq represents a major advancement in the study of RNA at the single-cell level. By providing a more detailed and accurate view of RNA isoforms and their variations, this technique opens up new possibilities for exploring the complexities of gene expression in health and disease, potentially leading to more targeted and effective therapies in the future.
Byrne A, Le D, Sereti K, Menon H, Vaidya S, Patel N, Lund J, Xavier-Magalhães A, Shi M, Liang Y, Sterne-Weiler T, Modrusan Z, Stephenson W. (2024) Single-cell long-read targeted sequencing reveals transcriptional variation in ovarian cancer. Nat Commun 15(1):6916. [article]
