Scientists have long believed that the human genome—the complete set of DNA in our bodies—contained a relatively fixed number of genes, each coding for a specific protein. However, recent discoveries have shown that there’s more to the story than we once thought. Researchers are now identifying previously hidden segments of DNA, known as small Open Reading Frames (smORFs), which can produce tiny proteins called microproteins. These microproteins may play crucial roles in various biological processes, but until now, finding them has been a significant challenge.
What Are smORFs and Microproteins?
An Open Reading Frame (ORF) is a sequence of DNA that has the potential to code for a protein. Traditionally, scientists focused on large ORFs, which produce the well-known proteins that perform most of the functions in our cells. However, smORFs are much smaller segments of DNA that also have the potential to produce proteins—just on a much smaller scale. The proteins produced by smORFs, known as microproteins, are tiny but may still have important roles in our cells.
The Challenge of Finding smORFs
One of the main challenges in discovering these smORFs has been the limitation of current research tools. Ribosome profiling (Ribo-Seq) is a technique used to identify which parts of the genome are being actively translated into proteins. While Ribo-Seq is great for finding large ORFs, it struggles with smORFs because these tiny sequences often overlap with other parts of the genome. This overlap creates confusion about which specific genomic location the translation activity is coming from, making it difficult to pinpoint the exact smORF responsible for producing a microprotein.
Introducing Rp3: A New Solution
To overcome these challenges, scientists at the Salk Institute for Biological Studies have developed a new method called Rp3. Rp3 is a pipeline—a series of steps that integrates different types of data to improve the accuracy of identifying smORFs. Specifically, Rp3 combines data from proteogenomics (the study of proteins and their encoding genes) with ribosome profiling. This combination allows researchers to more confidently detect and confirm the presence of microproteins that were previously missed by Ribo-Seq alone.
How Does Rp3 Work?
Rp3 enhances the ability to detect smORFs by addressing two major issues with traditional Ribo-Seq:
Multi-mapping Alignments: When a ribosome footprint (the part of the genome being read to make a protein) matches multiple places in the genome, traditional methods struggle to determine which location is correct. Rp3 can better handle these overlapping regions, providing clearer evidence of where translation is happening.
Proteomics Detection: While Ribo-Seq shows where translation occurs, it doesn’t directly identify the resulting proteins. Rp3 integrates proteogenomic data, which directly measures proteins, to confirm the existence of microproteins.
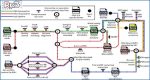
Workflow to reanalyze and overlay datasets
At the start of the workflow, RNA-Seq and Ribo-seq read undergo the same initial steps, starting with quality control and adapter trimming, followed by the alignment to the genome with STAR. Subsequently (I), the aligned RNA-Seq reads are used to assemble the transcriptome with StringTie, and this assembly is translated to the three-reading frames to predict the whole coding potential of that transcriptome, resulting in the three-frame translated (3FT) database (II) The aligned Ribo-seq reads are then used four times in the pipeline. First, they are used to score the ORFs from the transcriptome with both PRICE and Ribocode. Then, they are used as input for RibORF in step III. The reads are used one last time in step VI. (III) The alignments are scored against the 3FT database using RibORF, resulting in a fasta file containing Ribo-seq smORFs. A reference proteome is then appended to this fasta file to generate a custom database to check the mass spectrometry (MS) coverage for the Ribo-Seq smORFs. Similarly, the reference proteome is appended to the three-frame translated database, now without the Ribo-Seq smORFs, but with every predicted protein, which is the start of the proteogenomics pipeline. (IV) mzML files containing fragmentation spectra from MS experiments are searched against both databases using MSFragger, whose results are filtered with Percolator to obtain an FDR of 1%. This yields two subsets of results (V), Ribo-Seq smORFs covered by both Ribo-seq and MS evidence, and proteogenomics-derived smORFs (Rp3 smORFs), covered by MS evidence alone. The results from the first search are appended to the reference proteome and searched again with MSFragger, now with a reduced database instead of the 3FT. The search results are used as input for MSBooster to predict retention times and then fed to Percolator to assess the FDR. The Rp3 smORF-containing transcripts are located and (VI) the Ribo-seq reads containing secondary alignments are mapped to them using featureCounts (VII): once with default settings, to obtain Rp3 smORFs covered by Ribo-seq reads, and then allowing ambiguous and multi-mapping reads to be included during read counting, resulting in another subset of Rp3 smORFs covered by ambiguous and/or multi-mapped reads.
The development of Rp3 is significant because it opens up a new frontier in our understanding of the human genome. By revealing previously hidden smORFs and their microproteins, Rp3 allows scientists to explore new areas of biology that were once invisible. These microproteins could be involved in crucial processes such as cell signaling, metabolism, or disease progression, making them potential targets for new therapies and treatments.
Conclusion
The discovery of smORFs and microproteins represents a major expansion of the protein-coding genome. With the help of advanced tools like Rp3, researchers are now able to uncover these hidden elements of our DNA, providing new insights into how our cells function and how diseases might develop. As our understanding of the genome continues to grow, so too does the potential for innovative medical breakthroughs, making this an exciting time in the field of genetics and molecular biology.
Availability – https://github.com/Eduardo-vsouza/rp3
Vieira de Souza E, L Bookout A, Barnes CA, Miller B, Machado P, Basso LA, Bizarro CV, Saghatelian A. (2024) Rp3: Ribosome profiling-assisted proteogenomics improves coverage and confidence during microprotein discovery. Nat Commun 15(1):6839. [article]
