Retinal degeneration in mammals, which leads to permanent vision loss, is a challenging condition because, unlike some animals, mammals cannot naturally regenerate their retinal cells. However, research has shown that certain non-mammalian vertebrates, such as fish and amphibians, can regenerate retinal neurons through a specialized process involving Muller glia (MG), a type of supportive cell in the retina. This discovery has inspired scientists to explore ways to trigger similar regeneration in mammals, which could potentially lead to treatments for vision loss.
researchers at the University of Washington have made significant progress by stimulating adult mouse MG to regenerate functional neurons using a genetic approach. They did this by introducing a gene that activates a protein called Ascl1, a proneural transcription factor, which encourages the MG cells to take on a role similar to the neurons they support. While this method successfully generated new neurons in the retina, it was not highly effective. To improve this process, the researchers turned to a new technique to identify additional compounds that could boost regeneration.
They used a high-tech method called sci-Plex, which allows scientists to analyze thousands of single-cell samples at once using RNA sequencing (RNA-seq). This approach enables the team to look at the molecular activity within individual cells, offering a detailed view of how various compounds affect the regeneration process. In their study, the researchers screened a library of 92 different compounds and identified two that were particularly effective at promoting the growth of new neurons in the retina.
sci-Plex captures the in vitro temporal dynamics of neurogenic reprogramming from Muller glia (MG).
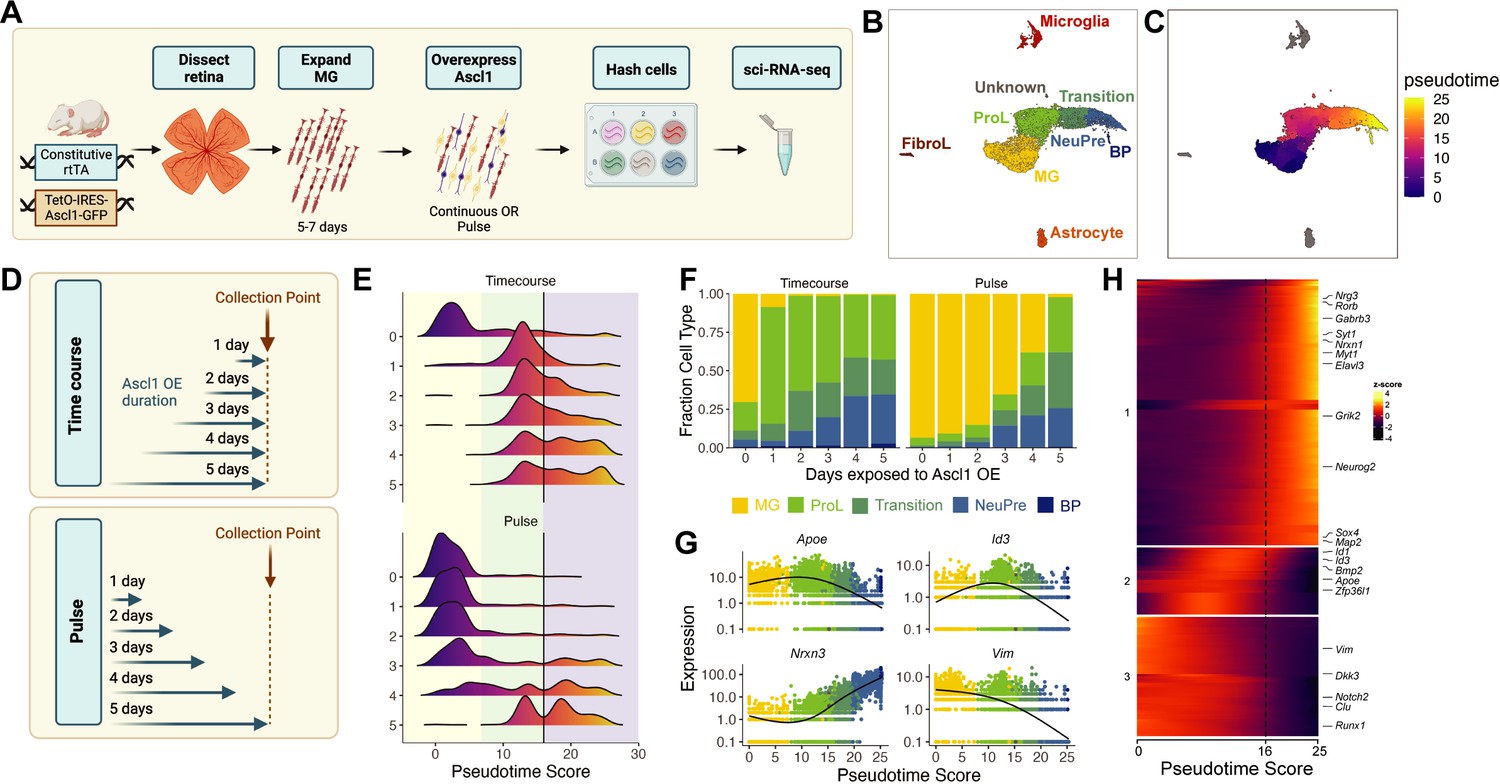
(A) Schematic of the sci-Plex experimental design for assaying reprogramming MG isolated from P11 mice. (B) Combined UMAP of cells from the Timecourse and Pulse experiments. Cells are colored by cell type. (C) UMAP displaying the pseudotime scores that were calculated for the MG to Bipolar trajectory. (D) Schematic depicting the timing and duration of Ascl1 OE in the Timecourse (n = 2 wells per condition) and Pulse (n = 3 wells per condition) experiments. (E) Histograms displaying the frequency of cells with each pseudotime score across Ascl1 OE conditions. The vertical black lines are at pseudotime score 16. The yellow region corresponds to the MG, the green region corresponds to the ProL cells, and the purple region reflects cells from the Transition to BP cell state. (F) Stacked bar plot of the cell type composition across all Ascl1 OE conditions. The colors represent the cell types as indicated. Only the MG and MG-derived cell types are included. (G) Gene expression plots along pseudotime for genes of interest. Each point represents an individual cell’s expression of the indicated gene. The cells are colored by cell type as in F. (H) Gene expression heatmap for the top 250 DEGs as assessed across a pseudotime score of 10–20. The dashed line is at pseudotime score 16. All cells with a non-infinite pseudotime score are ordered by pseudotime score along the x-axis. Genes were clustered by k-means into three clusters.
These findings are an exciting step forward. By using high-throughput single-cell profiling, the scientists were able to quickly and accurately identify molecules that encourage retinal regeneration. This technology could help uncover new pathways and molecules that promote neurogenesis, or the creation of new neurons, which could eventually be used to treat retinal diseases in humans and restore vision.
