Defining condensate-forming proteins
In order to develop a model to identify condensate-forming proteins, we assembled a ground truth dataset for H. sapiens, that has the most experimentally studied condensates of all organisms, to date. Since we aimed at developing a binary classifier, we considered two classes of proteins: (1) proteins involved in condensates (positive dataset) and (2) proteins not involved in condensates (negative dataset) (Fig. 1a). The positive dataset was constructed from a semi-manually curated dataset of biomolecular condensates and their respective proteins, called CD-CODE (CrowDsourcing COndensate Database and Encyclopedia), developed by our labs34. CD-CODE compiles information from primary literature and from four widely used databases of LLPS proteins25,26,27,28.
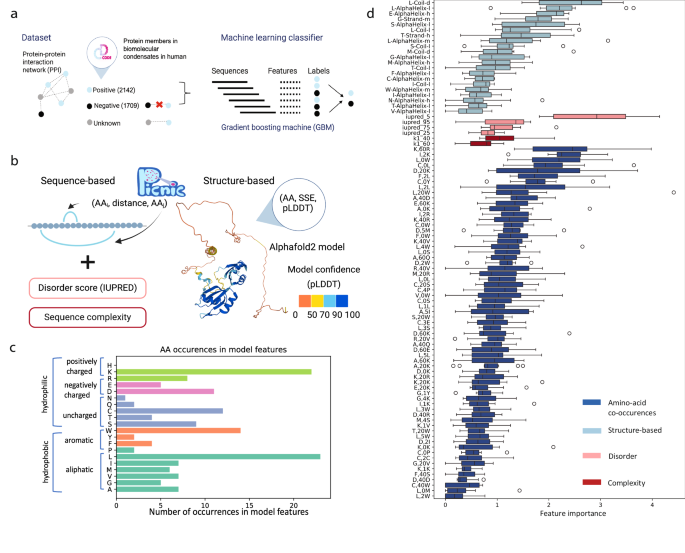
a In order to construct a training dataset, we annotated the known condensate-forming proteins from CD-CODE34 (positive dataset, members of biomolecular condensates) on the protein-protein interaction (PPI) network, and we excluded their first connections (proteins having interactions with condensate proteins). The remaining proteins comprised the negative dataset. Gradient boosting machine was used to distinguish two classes of proteins: members of biomolecular condensates and proteins that are not involved in any type of biomolecular condensate. b Sequence, structure and function-based features of PICNIC. Sequence-based features included sequence complexity, disorder score (IUPred), and features based on amino acid co-occurrences. Structure-based features based on AlphaFold2 models included the pLDDT score, a per-residue measure of local confidence on a scale from 0 to 100 (colored on the structure). We annotated the secondary structure (SSE) based on 3D protein structures using STRIDE and all possible triads in the form (AA, SSE, pLDDT) were calculated. c Amino acid occurrences in the features of PICNIC model show that Leucine and Lysine contribute most to the model predictions. d Feature importance of PICNIC is consistent across different folds (N = 10). The boxes show the quartiles of the dataset, where first black horizontal line of the rectangle shape is first quartile or 25% the second black horizontal line is the second quartile or median, the third black horizontal line is third quartile or 75%. The whiskers extend to points that lie within 1.5 IQRs (interquartile range) of the lower and upper quartile, the outliers are displayed as circles. Features constitute four groups: based on AlphaFold2structures (light blue), disorder (pink), complexity (dark red) and amino acid co-occurences (blue). Source data are provided as a Source Data file.
Building the negative dataset is a complicated task as there is no publicly available resource that reports proteins that do not form condensates. Additionally, condensates may form only under specific conditions35. Here, we defined the negative dataset based on protein-protein interaction network (InWeb database36 for human proteins). We excluded all proteins that have direct connections with known condensate proteins. We reasoned that these proteins are potential condensate members that have not yet been studied. The remaining proteins comprised the negative dataset (Fig. 1a). Of course, this procedure doesn’t guarantee the absence of condensate proteins among the negative dataset (false negatives). But exclusion of proteins that directly interact with proteins that were reported as members of synthetic or biomolecular condensates is lowering the probability of mixing positive and negative data. Overall, our non-redundant dataset (filtered by 50% sequence identity) contained 2142 positive and 1709 negative human proteins, which were divided by 4:1 ratio into training and test datasets.
PICNIC identifies sequence- and structure-determinants of condensate formation
We hypothesized that the ability to form condensates is encoded in the proteins’ sequence and structure, and developed a machine learning classifier called PICNIC (Proteins Involved in CoNdensates In Cell) based on sequence-distance and structure-based features derived from AlphaFold2 models (Fig. 1b), in total 65 sequence-distance-based and 21 structure-based features.
It was shown that many proteins involved in condensates harbor intrinsically disordered regions (IDRs) and low-complexity sequences. IDRs, due to their inherent flexibility, multi-valency and ability to sample multiple conformations, are adept at a wide array of binding-related functions including molecular assemblies9,37,38. Therefore, we also tested several metrics of disorder and sequence complexity as features (see Methods). Our final model contained several features related to disorder, such as IUPred scores33, that have a feature importance of 0.5–3%.
Although the presence of highly disordered residues is among the most important features (Fig. 1d, pink), it is not a prerequisite for a protein to have long disordered domains to be a member of a condensate(s). This is supported by the observation, that the proportion of known condensate-forming proteins with no disordered regions in the human proteome is 21% (disordered regions <10aa. Fig. S1), while 33% of all human proteins have no disordered regions. For example, Human protein Guanine nucleotide exchange factor C9orf72 is a driver protein in stress granules; Speckle-type POZ protein is a driver in nuclear speckle and SPOP/DAXX body. Both proteins consist of ordered domains that were experimentally determined by electron microscopy and X-ray crystallography, respectively (Fig. S1, PDB ids 6LT0 ad 3HU6). Thus, both the analysis of experimentally verified condensates and the selected features by our model suggest that disorder is not a necessity for condensate-forming proteins.
Along with overall sequence complexity and disorder scores of a protein, the secondary structure of individual residue types was also found to be important. We used the confidence score of the AlphaFold2 model prediction, the pLDDT score, that was shown to correlate with sequence disorder39. We represented the occurrence of an amino acid (AA) in a given secondary structure element (SSE) with a given model confidence as a triad (AA-SSE-pLDDT).
As amino acid composition bias and patterning of charges were shown to impact the ability of proteins to form condensates30,31,32, we developed features that represent short and long range co-occurrences of amino acids in the protein sequence. We represent co-occurrence of amino-acids in the protein sequence within a distance (number of amino acids in linear sequence) by triads (AA1, distance, AA2). After feature selection, the long-range distance between charged amino acids, e.g. Lysine and Arginine (K,60, R) and Aspartic acid and Lysine (D,20,K), and short-range distance of Leucine and hydrophobic amino acids (e.g. L,0,W; F,2,L; L,2,L), and the distance between Cysteine and hydrophobic amino acids were shown to be the most important features (Fig. 1d). Among the amino acids, Lysine and Leucine amino acids contribute the most to the model (Fig. 1c).
PICNIC accurately identifies proteins involved in biomolecular condensate formation
Several data-driven predictors were developed in the last few years, that aim to predict proteins involved in LLPS from protein sequence alone or from sequence and experimental data, such as microscopy images40. Here, we compared the performance of PICNIC to sequence based predictors, PSAP23, DeePhase41 and the general model of PhaSePred29 (PdPS-8fea based on 8 features) (Fig. 2).
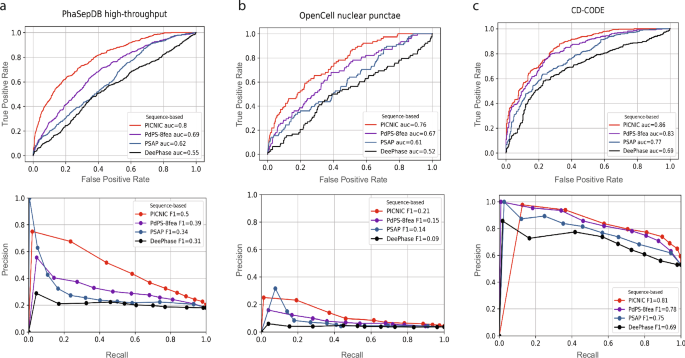
Comparison of sequence-based predictors (PICNIC, PdPS-8fea, PSAP, and DeePhase) of condensate proteins using different metrics. a Test dataset from PhaSepDB high-throughput retrieved from29 (441 positive and 1998 negative examples, excluding proteins that were part of the PICNIC training set), (b) test dataset from OpenCell42 (78 positive and 1998 negative examples excluding proteins that were part of the PICNIC training set), (c) test dataset from the current study based on CD-CODE34 (338 positive and 299 negative examples, i.e proteins that were not part of the PICNIC training set). PICNIC outperforms sequence-based predictors even on the test set that includes training data of previously published predictors, that may inflate their performance. Source data are provided as a Source Data file.
We compared the performance of tools on three different datasets: (1) test dataset from the recently published PhaSePred methods29; (2) proteins forming nuclear puncta defined by the OpenCell project42; (3) test dataset generated from CD-CODE34 (see Methods, Dataset S1). Although the CD-CODE test data is not independent and was partially used by existing predictors during their training process, PICNIC has superior performance with a maximum F1-score of 0.81 (Fig. 2c).
To further validate our model, we used microscopy images from Human Protein Atlas (HPA) where fluorescently labeled proteins were imaged and their cellular localization was determined43. Specifically, three types of cellular localizations were screened: nucleolus, centrosome and nuclear speckle. We filtered the list of proteins from HPA that were already in our training set that resulted in 484 proteins with known localization. Overall, PICNIC scores were higher for the proteins from HPA than for proteins without known localization (Fig. S4). 69% of proteins mapped from HPA (with exclusion of the proteins from the training dataset) have a PICNIC score greater than 0.5, meaning that PICNIC correctly identified them as members of biomolecular condensates. It should be noted that HPA doesn’t report if a protein does not belong to given condensate (negative examples). Therefore, this dataset can be used only to check model sensitivity (recall, what fraction of true condensate forming proteins were predicted correctly), but not model precision (what fraction of positive predictions are actually true positives).
PICNIC is robust in identifying small sequence perturbations that impact condensate formation
A challenging task for a computational predictor is to be sensitive to small sequence perturbations that can impact condensate formation. To test if PICNIC can distinguish similar sequences with altered condensate forming properties, we considered the synuclein family, that comprises three paralogs in human. Although they have similar sequences (60–70% identity, Fig. 3a, c) and structures as predicted by AlphaFold2 (Fig. 3b), only α- and γ-synuclein form condensates in vivo, and only α-synuclein phase separates in vitro. Specifically, FITC-labeled β-synuclein, which lacks the characteristic NAC region of α-synuclein, does not phase separate at high concentrations (200 μM) and under crowding conditions (10% [weight/volume] PEG), whereas FITC-labeled α-synuclein forms condensates under the same conditions22,44,45. While α- and γ-synuclein can form amyloid-like fibers, β-synuclein does not45,46. Moreover, α- and γ-synuclein are part of biomolecular condensates: α-synuclein is reported to be the member of synaptic vesicle pool condensate46, γ-synuclein is a member of the centrosome47, but β-synuclein has not been found in any biomolecular condensates yet. PICNIC is the only method tested here which accurately predicts the in vivo condensate-forming ability of the synuclein family (Fig. 3d). Other methods either give the same score for all three paralogs and/or do not predict the correct tendency of condensate formation in vivo. The features that stand out in β-synuclein and are absent in the most important features of α- and γ− synuclein (I-Alpha helix-l, F-Alpha helix-l) are connected to hydrophobic amino acids, being part of alpha-helix with low pLDDT score (Fig. S16). Thus, the structural changes involving the alpha helix are likely to drive the signal. We surmise that PICNIC is sensitive to structural rearrangements of proteins, and hypothesize that the bending of alpha-helix in β-synuclein potentially hinders the protein’s ability to form condensate as highlighted on the structure (Fig. S16).
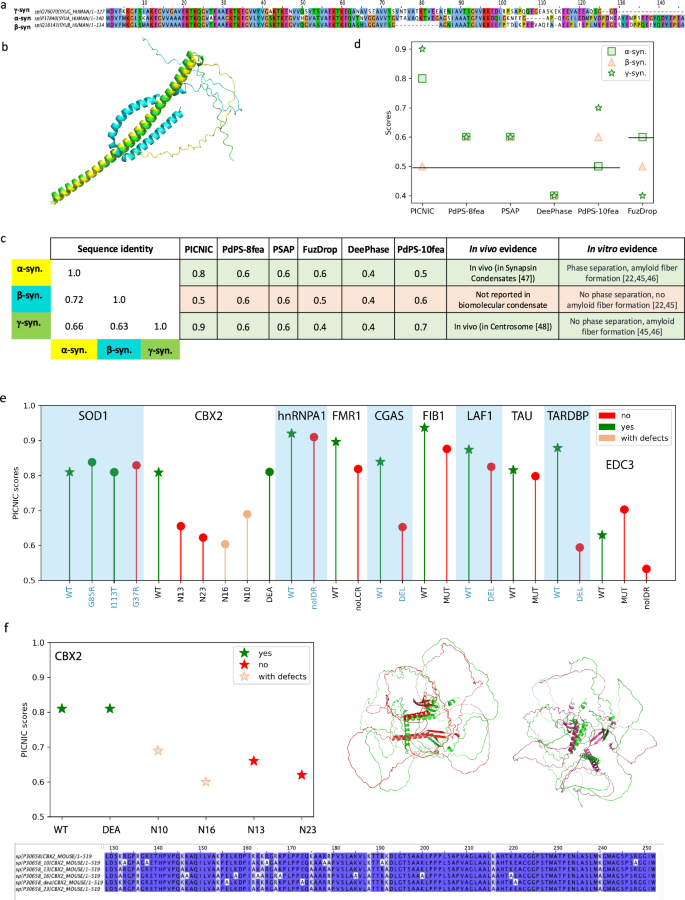
a The three paralogs of the synuclein family in human share high sequence identity as depicted in the multiple sequence alignment. b Structural models for α-synuclein (yellow), β-synuclein (cyan) and γ-synuclein (green), predicted by AlphaFold2 reveal that β-synuclein has a bent structure. c Despite the high sequence similarity, only α- and γ-synuclein are part of biomolecular condensates, while β-synuclein has not been found in any biomolecular condensates yet and was shown not to phase separate in vitro. d Comparison of prediction scores of different tools in identifying condensate forming (α and γ, green) and non-condensate forming paralog (β, red). PICNIC accurately predicts the condensate-forming ability of the synuclein family, and ranks β-synuclein the lowest, while other tools give equivalent scores to all paralogs or fail to identify the right trend. Vertical lines indicate the threshold used by the various methods to classify condensate-forming proteins. e PICNIC scores of WT (shown as stars) and mutant sequences assembled from the literature (Table S1). f Example of PICNIC’s performance on the mutated sequences of CBX273. Whereas the scores for the canonical sequence and mutant CBX2_DEA, that both form condensates (green stars) are high, the score decreases for the mutants with reduced ability to condensate (empty red stars) (CBX2_N10) and (CBX2_N16), and for the mutants CBX2_N13 and CBX2_N23 that do not form condensates (red stars). Sequence alignment of the canonical sequence of CBX2 and the mutated sequences studied here. On the left panel the structural alignment between CBX2 (green) and CBX2_N13 (red), as well as CBX2 (green) and CBX2_N23 (purple) points out that even with preserved SSE, their 3D orientation affects the proteins’ property to condensate. Source data are provided as a Source Data file.
This encouraged us to further test, if PICNIC can predict the impact of mutations, i.e. substitutions and deletions. We assembled a dataset of sequence perturbations that impact the ability of a protein to form condensates described in the literature. We excluded the most commonly used phase-separating proteins such as FUS because they are used as a model for many algorithms, and the performance of these proteins is heavily biased. To this end, our dataset comprised of proteins with 27 single mutations and deletions in 10 proteins in total (Table S1, Dataset S5). PICNIC scores were consistently lower for mutant sequences that have a reduced or completely abolished ability to form condensates (Fig. 3e, f). However, the scores are still higher than the threshold 0.5. This indicates that the impact of mutations is better resolved when interpreted in relation to each other, since the overall ability of the protein to form condensates is encoded in features that are shared across the different mutants. Nevertheless, PICNIC can predict the relative ability of mutants to form condensates.
Experimental validation of predicted condensate-forming proteins
In order to experimentally validate our model, we decided to predict the condensate localization of poorly characterized human proteins and sought to validate their condensate-forming behavior inside living cells. To do so, we chose 24 proteins which: (1) cover diverse molecular functions spanning the entire central dogma of molecular biology and regulation of all major cellular bio-polymers for instance, nucleic acids, proteins and chromatin (Fig. S8), (2) represent the average sequence length of human proteins (i.e., around 350 amino acids) by having a range of 125 to 684 amino acids, (3) represent diverse 3D structures from ordered, alpha-helical, beta-stranded to highly disordered (Fig. 4b) and (4) are known to be involved in genetic diseases (AIMP1, CWC27, RP9, LMOD1) as well as host-pathogen interaction (IF2GL). Overall, the 24 proteins (Dataset S2), we chose for experimentally verifying and benchmarking PICNIC, represent global cellular functions and therefore are suitable to demonstrate how robust our machine learning model is in predicting condensate-forming proteins across entire proteomes.
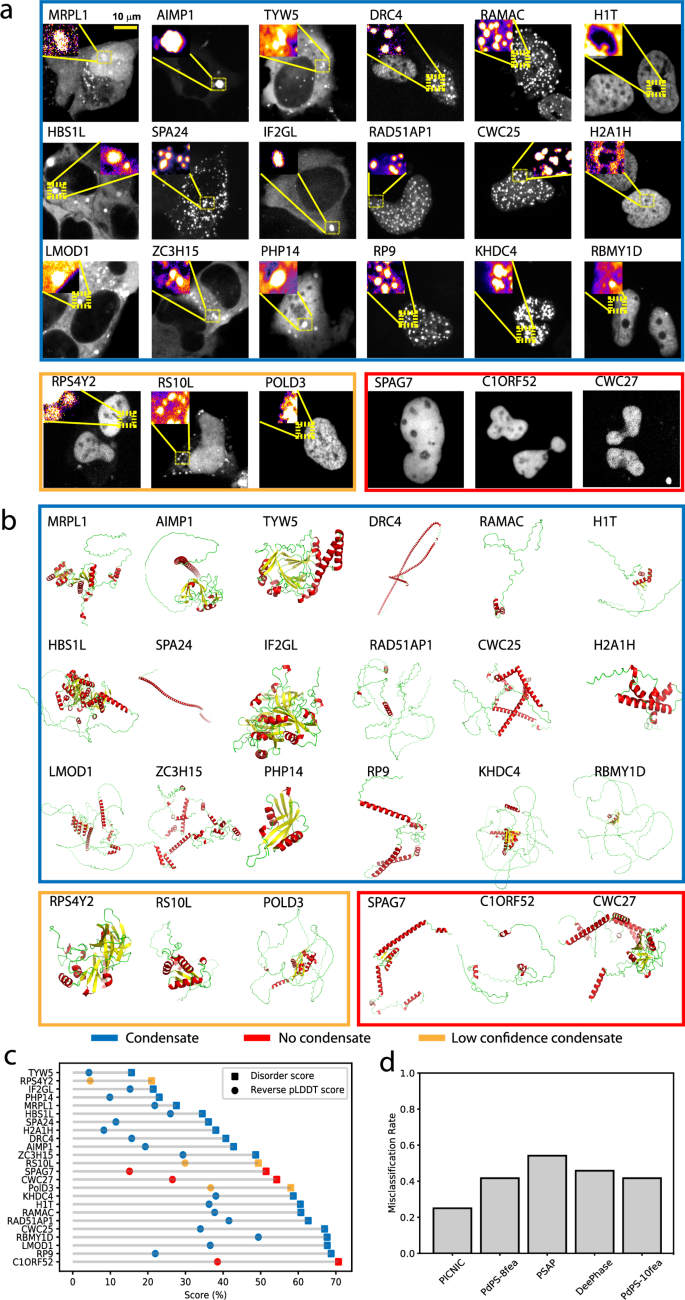
a Representative images of the U2OS cells expressing the tested proteins, N-terminally tagged with an iRFP fluorescent tag. Formation of mesoscale cellular condensates are highlighted in the inset. All images are scaled to the scale bar 10 μm (shown on upper left image). We found 21 out of the 24 tested proteins (87.5%) formed mesoscale foci without any stressors, while 3 proteins (C1ORF52, SPAG7 and CWC27, encircled in red) localized to the nucleoplasm without forming any detectable foci (foci were defined by exhibiting a fluorescent intensity ratio >1). Notice the presence of rim-like structures in case of H1T and H2A1H. Using size, shape and the fraction of cells forming mesoscale foci as a deciding characteristic, ~75% i.e., 18 proteins (encircled in blue) form high confidence condensates, and 3 proteins (encircled in orange) form low confidence condensates (foci with longest diameter <350 nm. Fig. S9). The experiments were repeated at least twice to ascertain the reproducibility of the results. More representative images are shown in Fig. S10 and the raw images are provided as Dataset S4. b Wide range of secondary structural motifs covered in the test proteins; AlphaFold2 structural models of the proteins are colored according to secondary structures. Notice the wide range of structural motifs, alpha-helical (red), beta stranded (yellow) to largely disordered (green) proteins (AlphaFold2 structures are provided as Dataset S6). c Disorder content (computed as mean IUPred score or reverse pLDDT score (1 – pLDDT)) of the tested protein does not correlate with the ability to form condensates. d Comparison of the predictions provided by sequence-based predictors (PICNIC, PdPS-8fea, PdPS-10fea, PSAP, and DeePhase) of protein condensates. PICNIC exhibits the lowest misclassification rate for the tested 24 proteins. Source data are provided as a Source Data file.
We cloned 24 transgenes and transfected them in U2OS cells expressing fluorescently labeled proteins (see Methods). Using fluorescent imaging, we found, that 21 out of the 24 tested proteins (87.5%) localized to mesoscale foci without any stressors, while 3 proteins (C1ORF52, SPAG7 and CWC27, encircled in red) localized to the nucleoplasm without forming any discernible mesoscale foci (Fig. 4a, Fig. S10). Foci were defined based on enrichment in fluorescent intensity, i.e., the intensity ratio inside relative to outside the foci is greater than one (Fig. S9a). In sum, only 3 tested proteins tested show close to no detectable foci, and 21 form foci (Fig. 4a).
In order to classify the observed foci as biomolecular condensates, we aimed to define quantitative characteristics and thresholds. We measured four simple characteristics from fluorescent microscopy images (Fig. S9): area and perimeter, informing on the size and the typical number of proteins in a foci; shape (roundness); number of foci per cell. Next, we decided on a threshold for these characteristics to aid a quantitative definition of condensates. We consider foci as condensates above the diameter of 350 nm (distance between two furthest pixels in one condensate), that is well above the diffraction limit. This would correspond to at least ~1 μm perimeter assuming a near round shape (Fig. S9c). Using back-of-the-envelope calculations, we can consider an average protein size as 10 nm3, then a 1 μm3 compartment can contain ca. 1 million protein molecules and a 500 nm3 compartment can contain 100,000 protein molecules. We note, that other super resolution techniques are required to characterize the size of the clusters of proteins below the diffraction limit.
Experiments confirm 87.5% of PICNIC predictions
By applying the above definition, in our dataset, 75 %, i.e., 18 proteins (encircled in blue) form high confidence condensates, 12.5 % i.e. 3 proteins (encircled in orange) form low confidence condensates (foci with perimeter <1 μm) while other 3 are not forming any condensates (encircled in red) and exhibit a fluorescent intensity ratio ~1. We observe most condensates to be round (Fig. S9d). The number of condensates per cell varies between 1 and 100 s depending on the protein of interest.
Next, we wanted to characterize the localization of the condensates that the proteins formed. We observed that 9 proteins (TYW5, SPA24, AIMP1, ZC3H15, IF2GL, LMOD1, RPS4Y2, DRC4, RS10L) localized to cytoplasmic bodies and 7 (RAD51AP1, KHDC4, CWC25, POLD3, RAMAC, DRC4, RP9) localize to nuclear bodies (Fig. 5a, Fig. S10).
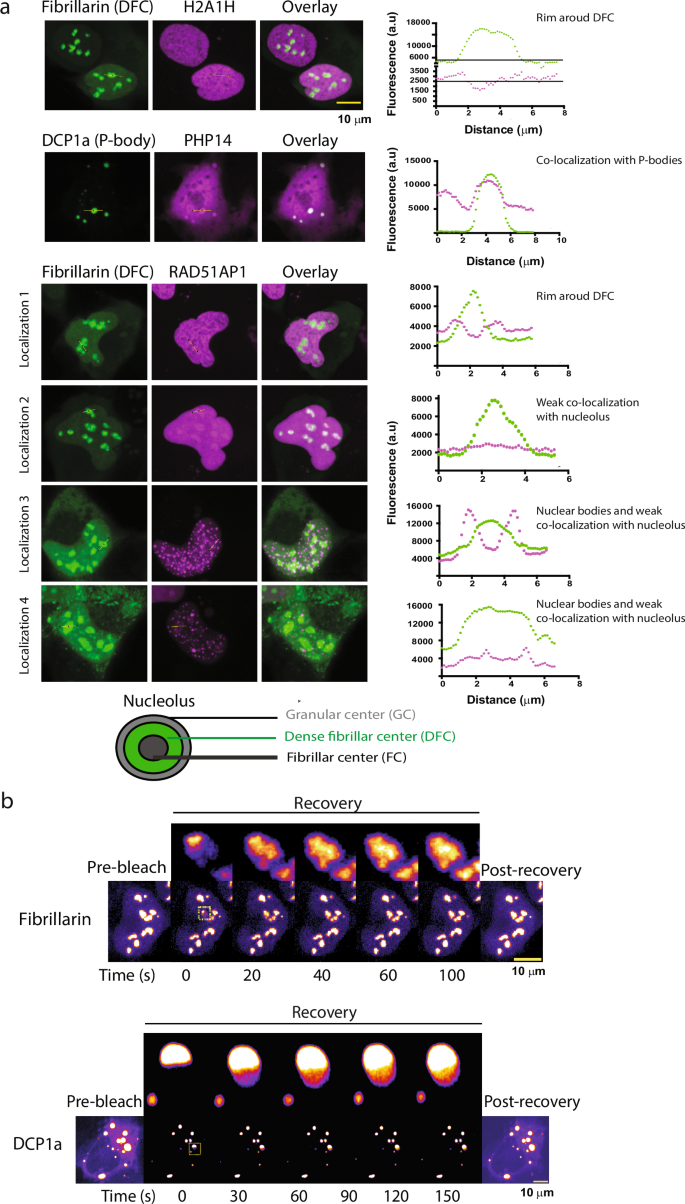
a Co-localization of the cellular condensate-forming proteins with well-characterized liquid-like cellular condensates as can be concluded from the fluorescence intensity profiles correlation with the marker protein fluorescence profiles (shown in green). While RAD51AP1 (in purple) localizes strongly around the Dense fibrillar center (DFC, in green) forming a rim like structure, H2A1H (in purple) show rather weak localization as a rim around the DFC (in green) showing sub-nucleolar localization specific to the outer Granular center (GC). See the cartoon representation of the nucleolar architecture. Further, PHP14 (purple) co-localizes with the DCP1a- labeled (green) processing bodies. RAD15-associated protein 1 (RAD51AP1) shows localization that varies from exclusive nuclear body like appearance (Localization 3, 4) to RAD51AP1 forming a rim around DFC part of the nucleolus (GC, Localization 1), abutting the nucleolus without forming a rim (Localization 3) and sub-nucleolar localization (Localization 4), as well as complete nucleolar localization suggesting an interesting role for this protein’s involvement on multiple nuclear-condensates (see also Fig. S10) possibly in a cell-cycle stage regulated manner. All images are scaled to the scale bar 10 μm (upper right corner) and available as Dataset S4. Co-localization experiments were repeated at least twice to ascertain the reproducibility of the results (see also Fig. S10). b FRAP assays showing the fast recovery dynamics consistent with the liquid-like nature of the P-bodies (upper panel) and the Nucleolus (lower panel). Fibrillarin and DCP1a FRAP recovery profile, inset highlighting the fast recovery dynamics of targeted P-body and the nucleolus. The scale bar is 10 μm for each. Source data are provided as a Source Data file.
Using co-localization experiments with known nucleolus (Fibrillarin) and processing body (P-body) (DCP1a) markers, we found that many proteins (H2A1H and RAD51AP1, H1T, MRPL1, RS10L, RPS4Y2, PolD3) can localize to the nucleolus at least in some cells (Fig. 5a, see also Fig. S10), a well-characterized liquid-like nuclear condensate (FRAP experiments in Fig. 5b), and PHP14 can localize to P-bodies, another well-characterized cytoplasmic condensate (Fig. 5a).
H2A1H (in purple) shows rather weak localization as a rim around the DFC (in green) showing sub-nucleolar localization specific to the outer Granular center (GC, see the cartoon representation of the nucleolar architecture). RAD15-associated protein 1 (RAD51AP1) shows multi-condensate localization that varies from exclusive nuclear bodies (Localization 3 and 4), to nuclear bodies abutting the nucleolus (Localization 3), and RAD51AP1 forming rim around DFC (GC, Localization 1) to sub-nucleolar localization (Localization 4) as well as complete but weak nucleolar localization (Localization 2) (Figs. 5a, S10) suggesting an interesting role for this protein’s involvement on multiple nuclear-condensates possibly in a cell-cycle stage regulated manner. The localization patterns of the condensate forming proteins are consistent with the wide-range of molecular functions that these proteins perform (Fig. S8). We also observed more than one type of foci in case of many proteins for e.g. DRC4: nuclear and cytoplasmic; MRPL1: nuclear and cytoplasmic bodies; RS10L: cytoplasmic bodies, filaments as well as nucleolar localization (Fig. S10).
In addition, FRAP recovery profiles for a subset of the condensate forming proteins (RAD51AP1, KHDC4, CWC25, RAMAC, DRC4, RP9, RBMY1D, TYW5) revealed, that 6 out of 8 tested proteins show very fast dynamics indicated by the recovery of the bleached foci, while two (TYW5 and RBMY1D) shows little to no recovery at the indicated time (Fig. 6).
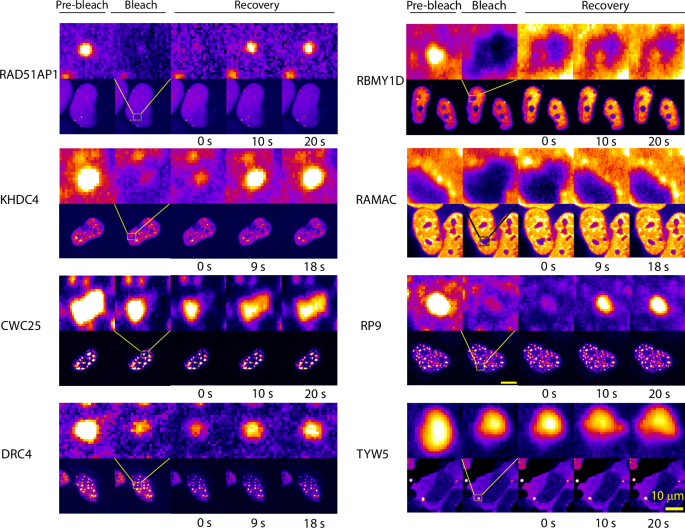
FRAP recovery profiles for the condensate forming proteins. 6 out of 8 tested proteins show very fast dynamics (at a time scale of 20 s, indicated) indicative of liquid-like nature, while two (RBMY1D and TYW5) show no recovery at the indicated time. All the images with indicated whole nuclei (lower panel for each protein) are scaled to the 10 μm scale bar shown in the bottom right. FRAP experiments were repeated at least twice to ascertain the reproducibility of the results. Images are provided as Dataset S4.
In sum, we do not see any correlation with disorder content of a protein and its ability to form condensates in our experimental dataset (Fig. 4c). Other popular tools to predict condensate proteins would fail to make correct predictions for many of these proteins as shown by the high misclassification rates (Fig. 4d). Overall, 87.5% of PICNIC predictions were found to be correct (misclassification rate is 25% for high confidence condensates and 12.5% if we include both high and low confidence condensates) in our experimental assays validating the model.
Proteome-wide predictions detect no correlation of predicted condensate proteome size with disorder content and organismal complexity
To demonstrate the generalizability of PICNIC, which was trained on human data, we tested its performance in identifying known condensate-forming proteins of other organisms. We screened the CD-CODE database34 to evaluate the fraction of proteins that were correctly identified as members of condensates by our predictor. PICNIC successfully predicted 72% of such proteins in mouse and 86% in Caenorhabditis elegans for example (Fig. 7a). Thus, PICNIC model is species-independent and is useful for different organisms to assess the ability of proteins to be involved in biomolecular condensates.
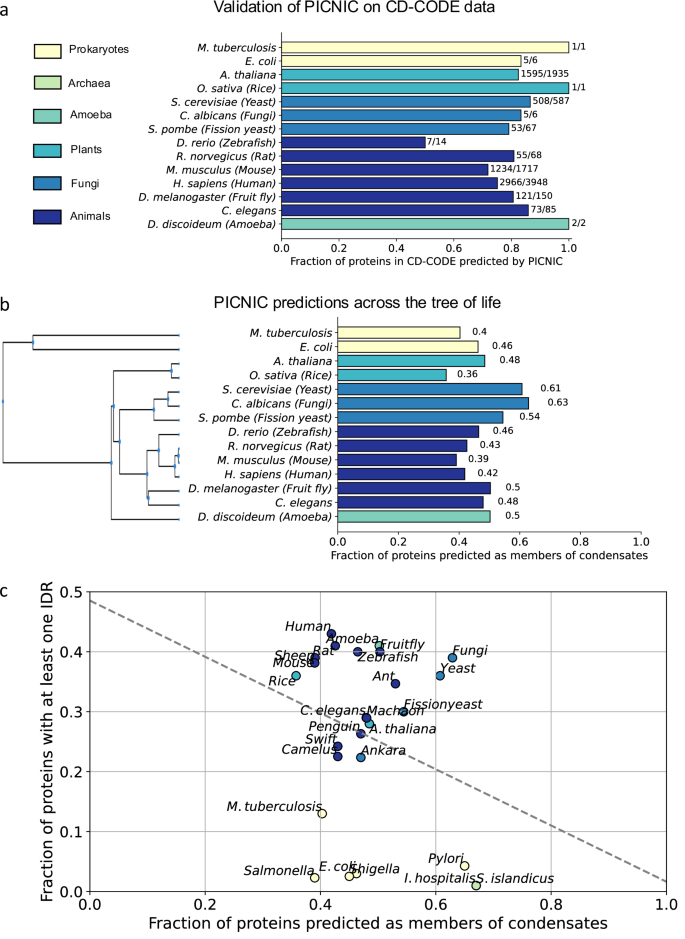
a PICNIC model is species-independent. We validated the PICNIC model on known condensate proteins from 14 different species (defined by CD-CODE)34. PICNIC correctly identified 70–100% of known condensate proteins of all species tested, except for zebrafish (50%). b Proteome-wide prediction of proteins in biomolecular condensates by PICNIC predictor. c Disorder content and fraction of condensate-forming proteins of a proteome are not correlated. The fraction of disordered proteins (proteins with at least one disordered region of > = 40 residues) in proteomes shows no correlation across 26 selected organisms from bacteria, archaea to mammals (Pearson R2 = 0.08). Source data are provided as a Source Data file.
To estimate the overall proteome fraction of condensate-proteins, we calculated PICNIC scores for 26 different organisms across the tree of life including bacteria, plants and fungi (Fig. 7). We selected 14 organisms that have already known condensate protein members that were experimentally verified in CD-CODE (we excluded organisms from further analysis where the number of known proteins is too small to compute statistics on the performance: Danio rerio (N = 14), Dictyostelium discoideum (N = 2), Escherichia coli (N = 6), Mycobacterium tuberculosis (N = 1), Oryza sativa (N = 1), Candida albicans (N = 6)). PICNIC correctly identified 50–100% of the known condensate proteins across organisms in CD-CODE (Fig. 7a). Although PICNIC was trained on human data, it generalized well and is likely applicable to proteomes of other organisms.
Next, we performed proteome-wide assessment of condensate proteins across 26 organisms. We found that the proportion of the predicted condensate-forming proteome is 40–60%, and is similar across related organisms, e.g., 42% and 39% in human and mouse, respectively (Fig. 7b). Interestingly, while the fraction of disordered proteins increases with organismal complexity as shown before48,49, we found no correlation between fraction of predicted condensate proteins in a proteome and the disordered protein content (Fig. 7c) across the 26 species tested even when using different metrics to assess the disorder of a proteome (Fig. S14). For example, E. coli and H. sapiens have both ~40% of their proteome predicted to be involved in biomolecular condensates (Fig. 7b).
